CRISPR/Cas9系統(tǒng)自發(fā)現(xiàn)以來,得到快速發(fā)展已被廣泛應(yīng)用生命科學(xué)基礎(chǔ)研究、基因治療、動植物育種改良等領(lǐng)域【1】。基于Cas9切口酶(nCas9)與脫氨酶結(jié)構(gòu)域/糖基化酶(MPG或UNG)的融合成的堿基編輯器(ABE,CBE,gGBE,gTBE),可高效實現(xiàn)A-to-G,C-to-T,C-to-G,G-to-C/T,以及T-to-G/C的堿基替換,為糾正突變的疾病位點提供了精準(zhǔn)高效基因編輯工具【2-6】。然而由于Cas9的體積過大(1368個氨基酸),基于nCas9的堿基編輯器難以實現(xiàn)單個AAV(4.7kb) 的包裝遞送,極大限制了在體基因編輯的發(fā)展應(yīng)用。近年來一系列緊湊型的Cas9蛋白【7-9】、Cas12f系列同源物【10-14】、以及其祖先蛋白TnpB【15,16】被報道,由于編輯活性有限,或缺乏HNH結(jié)構(gòu)域而難以改造為缺口酶,都限制用于堿基編輯器的開發(fā)。2021年,張鋒團隊發(fā)現(xiàn)由IS200/IS605轉(zhuǎn)座子超家族編碼的IscB核酸酶,被認為Cas9的祖先蛋白,具有與Cas9相似的HNH和RuvC結(jié)構(gòu)域,且僅有約500個氨基酸(約SpCas9的1/3大小)【17,18】,具有開發(fā)成微型堿基編輯器的潛力。
2023年,楊輝團隊通過對OgeuIscB/ωRNA系統(tǒng)的工程化改造,開發(fā)出了高效的OgeuIscB變體(enOgeuIscB),并通過融合脫氨酶結(jié)構(gòu)域,開發(fā)出高效迷你型堿基編輯器(miBE),推動了DNA單堿基編輯領(lǐng)域進入迷你型的“新時代”,具有極大的臨床應(yīng)用潛力【19】。然而,IscB/ωRNA系統(tǒng)需要嚴格的6位堿基靶序列鄰近基序(TAM)來識別目標(biāo)DNA,識別位點有限。因此,開發(fā)靶標(biāo)識別范圍更廣的高效小型IscB堿基編輯器是十分必要。
2024年8月15日,輝大(上海)生物科技有限公司研發(fā)團隊、復(fù)旦大學(xué)附屬眼耳鼻喉科醫(yī)院黃錦海團隊和中科院腦科學(xué)與智能技術(shù)卓越創(chuàng)新中心楊輝團隊合作在Nature Chemical Biology上發(fā)表題為Engineered IscB–ωRNA system with expanded target range for base editing的研究論文。該研究通過宏基因組數(shù)據(jù)挖掘,鑒定出19種具有不同TAM范圍的新型IscB-ωRNA系統(tǒng);綜合RNA結(jié)構(gòu)優(yōu)化、蛋白質(zhì)工程化改造、流式細胞術(shù)、脫靶檢測等技術(shù)手段,成功獲得在人類細胞內(nèi)具有靶標(biāo)識別范圍廣、更高效編輯活性的IscB系統(tǒng)(IscB.m16*);通過融合脫氨酶結(jié)構(gòu)域,進一步開發(fā)出基于新型IscB的迷你型腺嘌呤和胞嘧啶堿基編輯器,并在哺乳動物細胞和小鼠疾病模型中包括SpCas9-BE無活性的疾病位點上均驗證了其強大的堿基編輯效率和廣泛的靶標(biāo)識別能力,為未來精準(zhǔn)基因治療臨床應(yīng)用提供了強有力支持。

研究人員首先從200GB的宏基因組數(shù)據(jù)庫中挖掘出19個未被表征的新型IscB系統(tǒng),采用細菌耗竭實驗鑒定相應(yīng)的TAM序列;進一步利用熒光報告系統(tǒng),篩選出10個具有真核細胞活性的IscB系統(tǒng),其中IscB.m16表現(xiàn)出最高的編輯活性。為提高IscB.m16系統(tǒng)的活性并拓寬TAM范11:33:58圍,研究人員對IscB.m16核酸酶進行RuvC結(jié)構(gòu)域的精氨酸掃描突變和TAM識別相關(guān)位點的飽和突變,以及對其ωRNA進行莖環(huán)截短和堿基替換的優(yōu)化改造。通過多輪迭代的高通量熒光報告系統(tǒng)篩選,最終獲得了編輯活性高和TAM范圍寬的IscB.m16變體(IscB.m16*,即IscB.m16RESH-ωRNA)。通過細菌耗竭TAM序列識別實驗發(fā)現(xiàn),相較于野生型IscB.m16的TAM位點MRNRAA擴展到NNNGNA。
(Credit: Nature Chemical Biology)
在此基礎(chǔ)上,研究人員構(gòu)建了迷你型腺嘌呤堿基編輯器(IscB.m16*-ABE)和胞嘧啶堿基編輯器(IscB.m16*-CBE)。在哺乳動物細胞中,IscB.m16*-ABE的堿基編輯效率與SpG-ABE效率相當(dāng),顯著高于已報道的enOgeuIscB-ABE且有更廣的TAM兼容性。在人源化人源化杜氏肌營養(yǎng)不良癥(DMD)小鼠疾病模型中,單AAV包裝的IscB.m16*-CBE經(jīng)注射至肌肉組織后,成功并高效的將小鼠肌纖維中dystrophin蛋白水平恢復(fù)至野生型小鼠的40%,為DMD患者提供了一種有希望的基因治療策略。
總的來說,該研究通過對新型IscB的挖掘和優(yōu)化改造,開發(fā)出靶向范圍更廣的高活性、高特異性的迷你型堿基編輯工具IscB.m16*-BE,在基于AAV的基因治療應(yīng)用中顯示出獨特的優(yōu)勢和巨大的潛力。
參考文獻:
1. Porto, E.M., Komor, A.C., Slaymaker, I.M., and Yeo, G.W. (2020). Base editing: advances and therapeutic opportunities. Nature Reviews Drug Discovery 19, 839–859. https://doi.org/10.1038/s41573-020-0084-6.
2. Komor, A.C., Kim, Y.B., Packer, M.S., Zuris, J.A., and Liu, D.R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. https://doi.org/10.1038/nature17946.
3. Gaudelli, N.M., Komor, A.C., Rees, H.A., Packer, M.S., Badran, A.H., Bryson, D.I., and Liu, D.R. (2017). Programmable base editing of A?T to G?C in genomic DNA without DNA cleavage. Nature 551, 464–471. https://doi.org/10.1038/nature24644.
4. Kurt, I.C., Zhou, R., Iyer, S., Garcia, S.P., Miller, B.R., Langner, L.M., Grünewald, J., and Joung, J.K. (2021). CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat Biotechnol 39, 41–46. https://doi.org/10.1038/s41587-020-0609-x.
5. Tong, H., Liu, N., Wei, Y., Zhou, Y., Li, Y., Wu, D., Jin, M., Cui, S., Li, H., Li, G., et al. (2023). Programmable deaminase-free base editors for G-to-Y conversion by engineered glycosylase. National Science Review, nwad143. https://doi.org/10.1093/nsr/nwad143.
6. Tong, H., Wang, H., Wang, X., Liu, N., Li, G., Wu, D., Li, Y., Jin, M., Li, H., Wei, Y., et al. (2024). Development of deaminase-free T-to-S base editor and C-to-G base editor by engineered human uracil DNA glycosylase. Nat Commun 15, 4897. https://doi.org/10.1038/s41467-024-49343-5.
7. Chen, S., Liu, Z., Xie, W., Yu, H., Lai, L., and Li, Z. (2022). Compact Cje3Cas9 for Efficient In Vivo Genome Editing and Adenine Base Editing. The CRISPR Journal 5, 472–486. https://doi.org/10.1089/crispr.2021.0143.
8. Davis, J.R., Wang, X., Witte, I.P., Huang, T.P., Levy, J.M., Raguram, A., Banskota, S., Seidah, N.G., Musunuru, K., and Liu, D.R. (2022). Efficient in vivo base editing via single adeno-associated viruses with size-optimized genomes encoding compact adenine base editors. Nat. Biomed. Eng 6, 1272–1283. https://doi.org/10.1038/s41551-022-00911-4.
9. Zhang, H., Bamidele, N., Liu, P., Ojelabi, O., Gao, X.D., Rodriguez, T., Cheng, H., Kelly, K., Watts, J.K., Xie, J., et al. (2022). Adenine Base Editing In Vivo with a Single Adeno-Associated Virus Vector. GEN Biotechnology 1, 285–299. https://doi.org/10.1089/genbio.2022.0015.
10. Hino, T., Omura, S.N., Nakagawa, R., Togashi, T., Takeda, S.N., Hiramoto, T., Tasaka, S., Hirano, H., Tokuyama, T., Uosaki, H., et al. (2023). An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis. Cell 186, 4920-4935.e23. https://doi.org/10.1016/j.cell.2023.08.031.
11. Kim, D.Y., Lee, J.M., Moon, S.B., Chin, H.J., Park, S., Lim, Y., Kim, D., Koo, T., Ko, J.-H., and Kim, Y.-S. (2022). Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat Biotechnol 40, 94–102. https://doi.org/10.1038/s41587-021-01009-z.
12. Kong, X., Zhang, H., Li, G., Wang, Z., Kong, X., Wang, L., Xue, M., Zhang, W., Wang, Y., Lin, J., et al. (2023). Engineered CRISPR-OsCas12f1 and RhCas12f1 with robust activities and expanded target range for genome editing. Nat Commun 14, 2046. https://doi.org/10.1038/s41467-023-37829-7.
13. Xu, X., Chemparathy, A., Zeng, L., Kempton, H.R., Shang, S., Nakamura, M., and Qi, L.S. (2021). Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. Molecular Cell 81, 4333-4345.e4. https://doi.org/10.1016/j.molcel.2021.08.008.
14. Wu, Z., Zhang, Y., Yu, H., Pan, D., Wang, Y., Wang, Y., Li, F., Liu, C., Nan, H., Chen, W., et al. (2021). Programmed genome editing by a miniature CRISPR-Cas12f nuclease. Nat Chem Biol 17, 1132–1138. https://doi.org/10.1038/s41589-021-00868-6.
15. Karvelis, T., Druteika, G., Bigelyte, G., Budre, K., Zedaveinyte, R., Silanskas, A., Kazlauskas, D., Venclovas, ?., and Siksnys, V. (2021). Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature 599, 692–696. https://doi.org/10.1038/s41586-021-04058-1.
16. Kim, D.Y., Chung, Y., Lee, Y., Jeong, D., Park, K.-H., Chin, H.J., Lee, J.M., Park, S., Ko, S., Ko, J.-H., et al. (2022). Hypercompact adenine base editors based on transposase B guided by engineered RNA. Nat Chem Biol 18, 1005–1013. https://doi.org/10.1038/s41589-022-01077-5.
17. Altae-Tran, H., Kannan, S., Demircioglu, F.E., Oshiro, R., Nety, S.P., McKay, L.J., Dlaki?, M., Inskeep, W.P., Makarova, K.S., Macrae, R.K., et al. (2021). The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science 374, 57–65. https://doi.org/10.1126/science.abj6856.
18. Schuler, G., Hu, C., and Ke, A. (2022). Structural basis for RNA-guided DNA cleavage by IscB-ωRNA and mechanistic comparison with Cas9. Science 376, 1476–1481. https://doi.org/10.1126/science.abq7220.
19. Han, D., Xiao, Q., Wang, Y., Zhang, H., Dong, X., Li, G., Kong, X., Wang, S., Song, J., Zhang, W., et al. (2023). Development of miniature base editors using engineered IscB nickase. Nat Methods. https://doi.org/10.1038/s41592-023-01898-9.
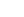




 17312606166
17312606166